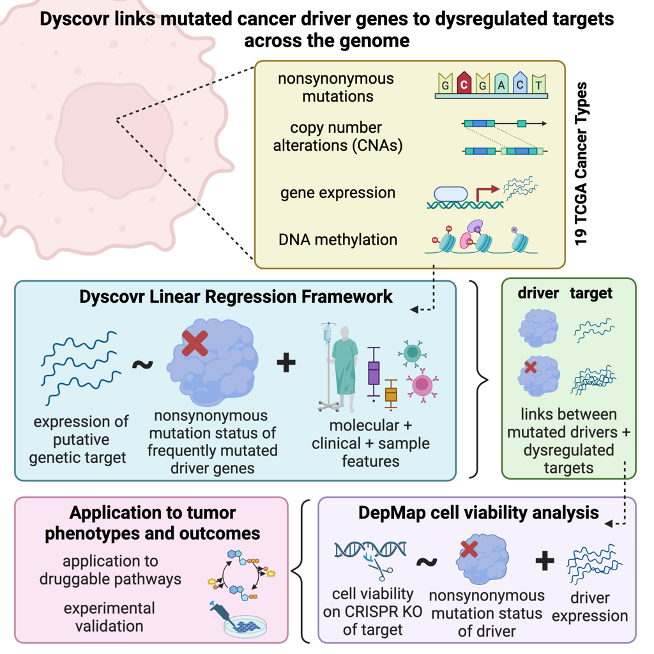
Search the output of Dyscovr, an integrative linear regression-based method developed by the Singh lab at Princeton University, by driver gene(s), putative target gene(s), and cancer type(s).
If you use Dyscovr's output in your own work, please cite as:
Geraghty, S., Boyer, J.A., Fazel-Zarandi, M., Arzouni, N., Ryseck, R., McBride, M., Parsons, L.R., Rabinowitz, J., Singh M. (2024). Integrative Computational Framework, Dyscovr, Links Mutated Driver Genes to Expression Dysregulation Across 19 Cancer Types. bioRxiv.